In the era of Precision Medicine, using gene expression,
DNA and other omics data to predict the response of cancer patients to Immuno-, Chemo- and targeted therapies is timely and important.
My team has developed Featuer selection and Machine Learning methods to reveal signatures for the prediction of response to
|
|
(1) Immune-Checkpoint inhibitors (ICIs), Atezolizumab, Nivozumab and pembrolizumab for patients with mUC, Renal Cell Carcinoma, and Melanoma.
(2) targeted- and chemo-therapies, e.g., Erlotinib and Cetuximab; see Yuan et al., (2023) for details.
|
|
Current projects including
|
|
(i) Analyzing more datasets in melanoma to identify robust signatures and prediction AUCs (of the ROC) in melanoma.
(ii) A collaboration with prof Yi-Chun Wu, NTU. To show circulating tumor cells (CTCs) of blood samples from patients with breast cancer can reveal mutation profiles, prognostic markers and signatures for the prediction of response to drug treatments,
chemotherapy or anti-HER2/anti-PD-L1 treatmen. Eventually to establish new indicators for prognosis and the prediction of drug response.
|
|
Projects
|
| (1) |
Machine Learning-based projects on precision medicine.
To find signatures for prediction of response of patients with Urothelial/Kidney cancer to immunotherapies.

(collaborate with Dr. Tai-Lung Cha, Director of National Institute of Cancer Research, NHRI) |
|
| (2) |
Identifying Rare Variants of gout in Taiwanese males via whole-genome sequencing.

(collaborate with Prof. Jan-Gowth Chang (China Medical Univ./Changhua Show Chwan Memorial Hospital)) |
|
| (3) |
Using deep learning (cNN) to identify cancer patients who can response to drugs.
|
|
| (4) |
Identifying (a) prognostic markers, (b) drug target and (c) key genes in colorectal/Lung/ Oral cancers.
(collaborated with profs. Pai-Chye Yang (National Taiwan Univ.); Joanne Chen (IBMS); Jan-Gowth Chang (CMU), and others)
Related publications:
Tiong et al. (2014), Hwang et al. (2015),
Chang et al. (2016), Wang et al. (2019). |
|
Before 2014
|
| • |
Using several kinds of genomic data to infer genetic interactions/pathway components. |
|
| • |
Using genetic interactions to develop novel cancer therapeutic agents.
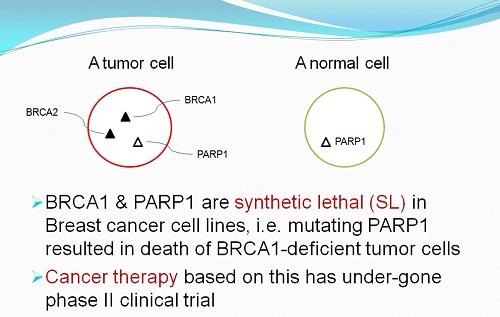
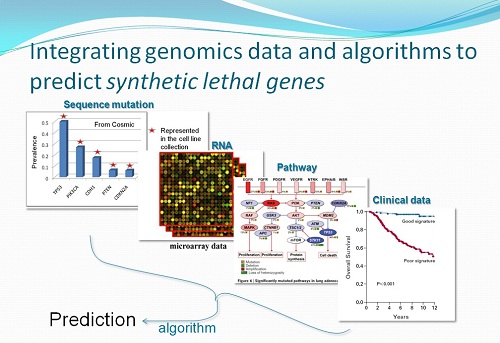
(with Prof. Jan-Gowth Chang(Kaoshung Medical Univ.), Dr. Konan Peck(IBMS, Academia Sinica), Jeou-Yuan Chen(IBMS, Academia Sinica)) |
|
| • |
Identification of the common regulators for hepatocellular carcinoma induced by hepatitis B virus X antigen in a mouse model
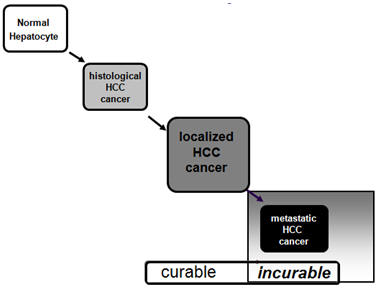
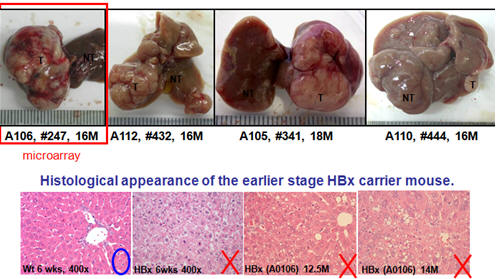
(with Dr. Cathy Yuh, NHRI) |
|
| • |
Epigenetics in yeast via ChIP-chip
and microarray data
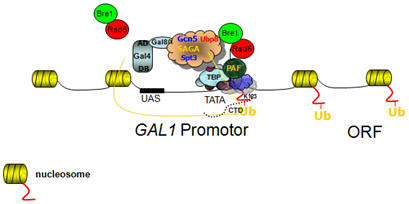
(with Dr. Cheng-Fu Kao, Inst. Cellular & Organ. Biology, AS) |
|
| • |
Predicting molecular
mechanism for Obesity
(with Profs. Jean-Daniel Zucker, Univ. Paris-13 and Karine Clement, INSERM, France) |
|
| • |
A Pattern Recognition Approach
to Infer time-lagged Genetic Interactions
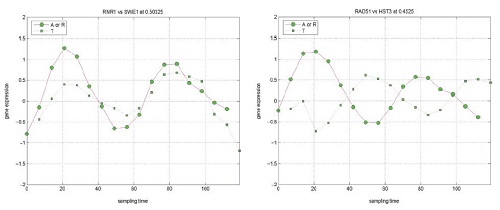
(with Prof.
Chung-Ming Chen, Inst. Biomedical Engineering, NTU) |
|
|
|